Many proteins are highly efficient biomolecular “nanomachines”. Just one example out of a myriad is bacteriorhodopsin (bR): it is a light-driven proton pump that transports protons out of the cell, a 10000-fold difference in proton concentration resulting in on each side of the membrane. The reverse proton flow serves the production of ATP using another biomolecular nanomachine, the FOF1-ATPase. Being a prominent member of the large family of G-protein-coupled seven-helix receptors the bR molecule consists of seven helices that span the membrane and feature an internal channel-like structure. There is growing evidence that so-called local area networks (WLANs) of hydrogen bonded water clusters, connected to amino acid side chains via hydrogen bonds, exist inside the channel. Again, water seems to play a key role in a living system- rather than being just an inert spectator (see also a related Feature Page in this Media Gallery).
This very role of water is one of the aspects that are in the focus of the Joint Research Group on “Interfacial Water” at the Universities of Bochum and Dortmund (DFG Forschergruppe 436 “Polymorphismus, Dynamik und Funktion von Wasser an molekularen Grenzflächen”).
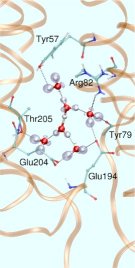
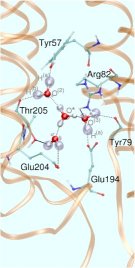
In the specific case of bR it is believed that such WLANs are at the very heart of storing, transporting, and releasing excess protons during its finely-tuned photocycle. This would imply that water - not only the bare protein - plays an active role in the function of this and probably many other such nanomachines. In particular, a rather large and hydrophilic cavity close to the extracellular membrane surface of bR, located in the lower third of the figure (see Fig. 1),
accomodates such a WLAN, consisting of several water molecules in the vicinity of Glu204 and Glu194, although the precise number of water molecules is not known. There is strong infrared spectroscopic evidence that a proton is stored in this WLAN just before it is released into the extracellular compartment, which therefore would be (part of) the so-called “proton release group”, XH. Thus, water might indeed be the “smallest biomolecule” (Philip Ball, Life's Matrix - A Biography of Water, 1999; H2O - Biographie des Wassers, 2001).
Clearly, it is currently not feasible to simulate the relevant system, that is the membrane protein bR embedded in a solvated membrane, using ab initio molecular dynamics. It is also clear that the usual harmonic approximation for calculating infrared spectra is not appropriate in view of the strongly anharmonic motion known to be present even in protonated water clusters in the gas phase (see also a related Feature Page in this Media Gallery). But thanks to advances in the coupling of quantum-mechanical and molecular mechanics approaches, the so-called “QM/MM” techniques, it has recently been possible to perform such simulations in the framework of Car-Parrinello molecular dynamics on sufficiently long time scales.
Only a small, but defintely the most important part of the system, the so-called “hot spot”, is treated with electronic structure methods, whereas all the rest is calculated using standard biomolecular force fields. In the present case the protonated water network in the lower third of the protein is taken into account quantum-mechanically, whereas the biomatrix comprising the protein and the membrane plus the solvent water is represented by classical mechanics. This allows one to compute the infrared spectra of the protonated WLAN, which is experimentally accessible via difference spectroscopy, beyond the harmonic approximation (i.e. from the Fourier transform of the dipole autocorrelation function).
But how to start such a simulation without even knowing the number of water molecules that contribute to this WLAN? It is known from proton migration in water according to the famous Grotthuss mechanism of structural diffusion that two protonation patterns might be preferred by the “wet proton” (see also a related Feature Page in this Media Gallery). One of them, the so-called “Zundel complex”, consists of two hydrogen bonded water molecules that share a proton, that is located right at the center of this peculiar hydrogen bond (see left picture in Fig. 3).
In the so-called “Eigen complex”, the excess proton is covalently attached to a water molecule thus forming a hydronium core, H3O+, which is solvated by three water molecules (see right picture in Fig. 3). These two archetypical protonated WLAN topologies have been embedded in the above-mentioned hydrophilic pocket as the QM part in a QM/MM setup of the entire system. The resulting infrared spectra turn out to be qualitatively very different. They are even complementary in the sense that the Zundel spectrum has its major spectral density below roughly 2000 cm-1 whereas the Eigen spectrum dominates the spectral range above about 2000 cm-1 Such calculations and the corresponding structural assignments will eventually allow one to uniquely determine the dynamical change in the protonation pattern of the excess protons as they get pumped through bR during its photocycle.
Further information and details:
- Hybrid QM/MM Car-Parinello biomolecular dynamics and analysis
- Gerald Mathias (Ruhr-Universität Bochum)
- Marcel Baer (Ruhr-Universität Bochum)
- Volker Kleinschmidt (Ruhr-Universität Bochum)
- Roger Rousseau (Ruhr-Universität Bochum, now SISSA Trieste)
- Udo Schmidt (Ruhr-Universität Bochum, now MPI Göttingen)
in collaboration with - Dominik Marx (Ruhr-Universität Bochum)
- Scientific visualization
- Nikos Doltsinis (Ruhr-Universität Bochum)
- Axel Kohlmeyer (Ruhr-Universität Bochum, now CMM at PENN)
- Additional information
- RUB Pressemitteilung
- There is a 10 minute animation DVD available: please contact theochem@theochem.rub.de
- Cover of the Bunsenmagazin from March 2004
- Wire-like topologies and flat free energy landscapes of water networks in bR:
G. Mathias and D. Marx,
Structures and spectral signatures of protonated water networks in bacteriorhodopsin,
Proc. Natl. Acad. Sci. USA 104, 6980 (2007) - Feature Article (in German) by Volkhard Helms: WLAN für die Zelle
- Computed infrared spectra of protonated water clusters in bR:
R. Rousseau, V. Kleinschmidt, U. W. Schmidt, and D. Marx,
Assigning Protonation Patterns in Water Networks in Bacteriorhodopsin Based on Computed IR Spectra,
Angew. Chem. Int. Ed. 43, 4804 (2004) - R. Rousseau, V. Kleinschmidt, U. W. Schmidt, and D. Marx,
Zuordnung der Protonierungsmuster von Wassernetzwerken in Bakteriorhodopsin mittels berechneter Infrarotspektren,
Angew. Chem. 116, 4908 (2004) - Structure and dynamics of protonated water clusters in bR:
R. Rousseau, V. Kleinschmidt, U. W. Schmidt, and D. Marx,
Modeling protonated water networks in bacteriorhodopsin,
Phys. Chem. Chem. Phys. 6, 1848 (2004) - Quantum correction factors for IR spectra of hydrogen bondend systems:
R. Ramírez, T. López-Ciudad, P. Kumar P, and D. Marx,
Quantum corrections to classical time-correlation functions:
Hydrogen bonding and anharmonic floppy modes,
J. Chem. Phys. 121, 3972 (2004) - Expert introduction into simulation techniques
- This web page
http://www.theochem.ruhr-uni-bochum.de/go/br.html